In vivo genome editing via CRISPR/Cas9 mediated homology-independent targeted integration
Authors
12-07-2016
12:00pm
PST
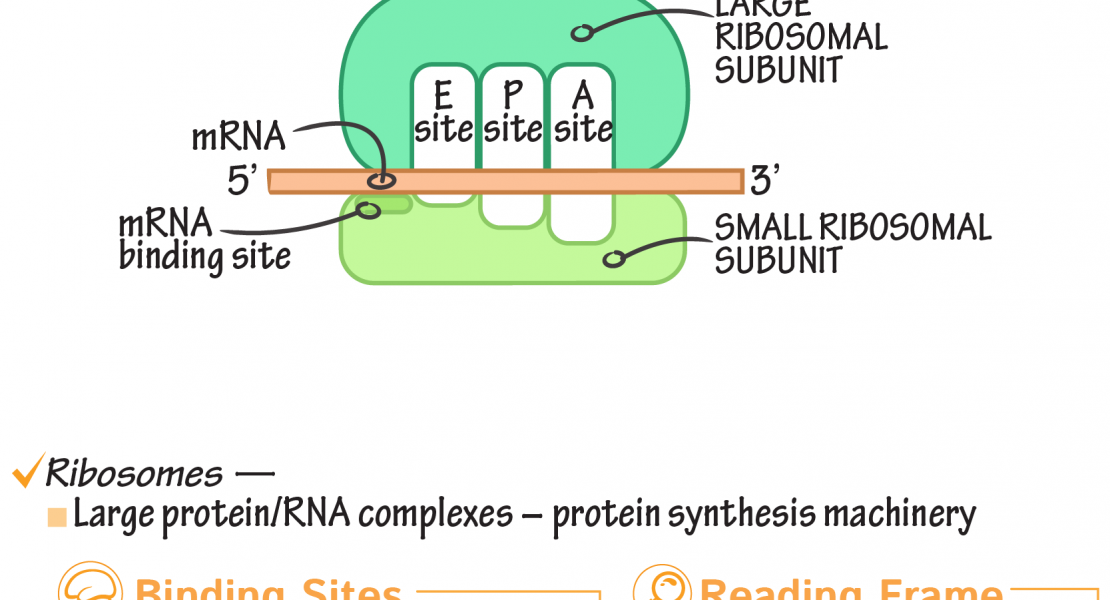
Abstract
The impact of RNA structures in coding sequences (CDS) within mRNAs is poorly understood. Here, we identify a novel and highly conserved mechanism of translational control involving RNA structures within coding sequences and the DEAD-box helicase Dhh1. Using yeast genetics and genome-wide ribosome profiling analyses, we show that this mechanism, initially derived from studies of the Brome Mosaic virus RNA genome, extends to yeast and human mRNAs highly enriched in membrane and secreted proteins. All Dhh1-dependent mRNAs, viral and cellular, share key common features. First, they contain long and highly structured CDSs, including a region located around nucleotide 70 after the translation initiation site; second, they are directly bound by Dhh1 with a specific binding distribution; and third, complementary experimental approaches suggest that they are activated by Dhh1 at the translation initiation step. Our results show that ribosome translocation is not the only unwinding force of CDS and uncover a novel layer of translational control that involves RNA helicases and RNA folding within CDS providing novel opportunities for regulation of membrane and secretome proteins.
