RNA Duplex Map in Living Cells Reveals Higher-Order Transcriptome Structure
Abstract
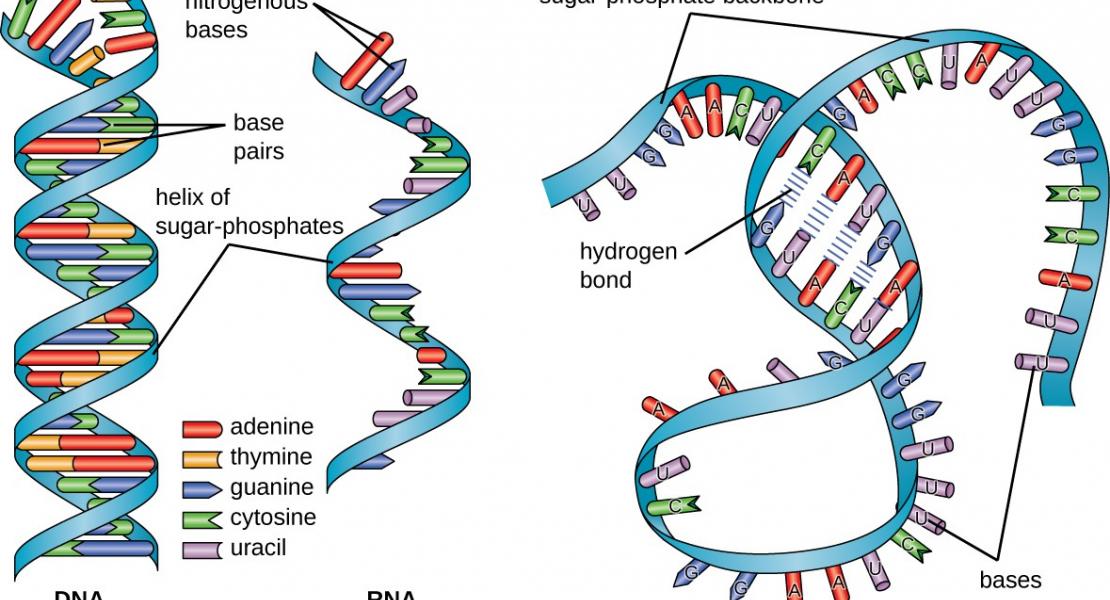
RNA has the intrinsic property to base pair, forming complex structures fundamental to its diverse functions. Here, we develop PARIS, a method based on reversible psoralen crosslinking for global mapping of RNA duplexes with near base-pair resolution in living cells. PARIS analysis in three human and mouse cell types reveals frequent long-range structures, higher-order architectures, and RNA-RNA interactions in trans across the transcriptome. PARIS determines base-pairing interactions on an individual-molecule level, revealing pervasive alternative conformations. We used PARIS-determined helices to guide phylogenetic analysis of RNA structures and discovered conserved long-range and alternative structures. XIST, a long noncoding RNA (lncRNA) essential for X chromosome inactivation, folds into evolutionarily conserved RNA structural domains that span many kilobases. XIST A-repeat forms complex inter-repeat duplexes that nucleate higher-order assembly of the key epigenetic silencing protein SPEN. PARIS is a generally applicable and versatile method that provides novel insights into the RNA structurome and interactome.
