Mechanism of eukaryotic RNA polymerase III transcription termination
Abstract
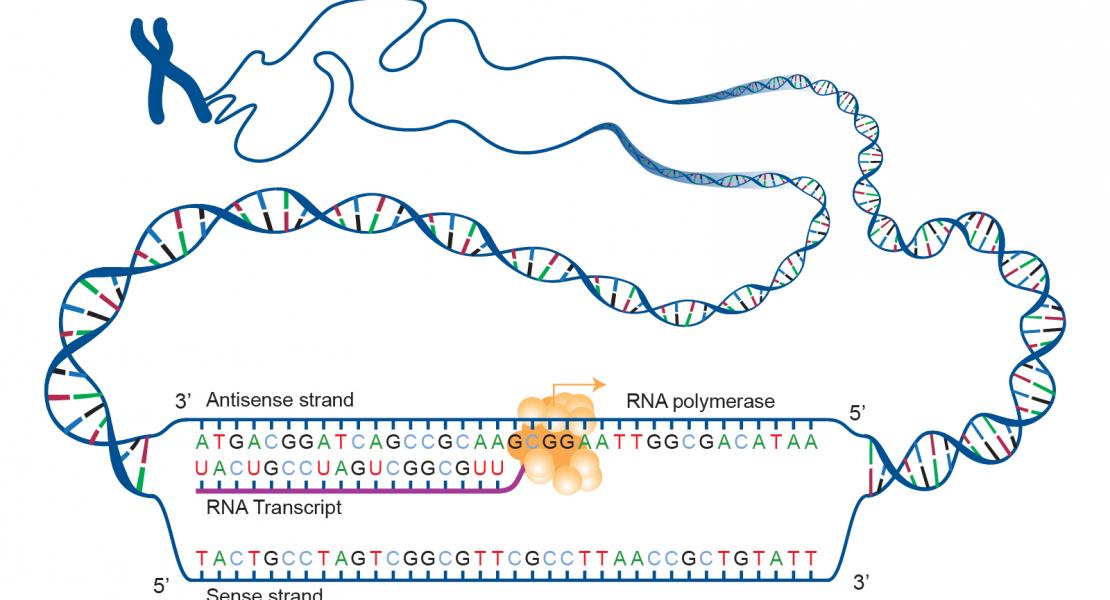
Gene expression in organisms involves many factors and is tightly controlled. Although much is known about the initial phase of transcription by RNA polymerase III (Pol III), the enzyme that synthesizes the majority of RNA molecules in eukaryotic cells, termination is poorly understood. Here, we show that the extensive structure of Pol III–synthesized transcripts dictates the release of elongation complexes at the end of genes. The poly-T termination signal, which does not cause termination in itself, causes catalytic inactivation and backtracking of Pol III, thus committing the enzyme to termination and transporting it to the nearest RNA secondary structure, which facilitates Pol III release. Similarity between termination mechanisms of Pol III and bacterial RNA polymerase suggests that hairpin-dependent termination may date back to the common ancestor of multisubunit RNA polymerases.
