Extensive transcriptional heterogeneity revealed by isoform profiling
Abstract
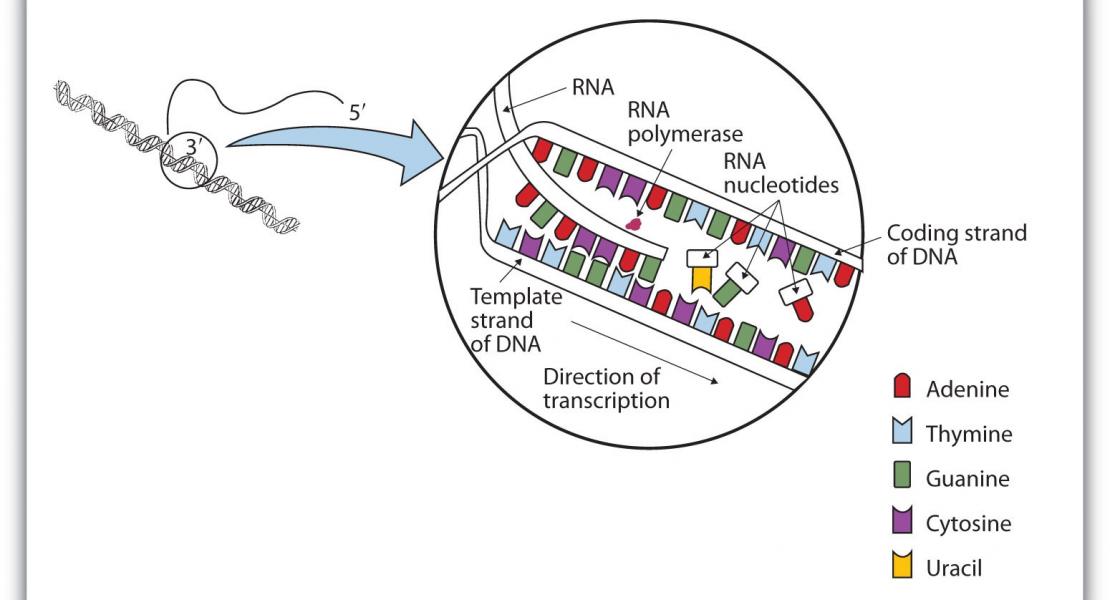
Transcript function is determined by sequence elements arranged on an individual RNA molecule. Variation in transcripts can affect messenger RNA stability, localization and translation1 , or produce truncated proteins that differ in localization2 or function3 . Given the existence of overlapping, variable transcript isoforms, determining the functional impact of the transcriptome requires identification of full-length transcripts, rather than just the genomic regions that are transcribed4,5. Here, by jointly determining both transcript ends for millions of RNA molecules, we reveal an extensive layer of isoform diversity previously hidden among overlapping RNA molecules. Variation in transcript boundaries seems to be the rule rather than the exception, even within a single population of yeast cells. Over 26 major transcript isoforms per protein-coding gene were expressed in yeast. Hundreds of short coding RNAs and truncated versions of proteins are concomitantly encoded by alternative transcript isoforms, increasing protein diversity. In addition, approximately 70% of genes express alternative isoforms that vary in post-transcriptional regulatory elements, and tandem genes frequently produce overlapping or even bicistronic transcripts. This extensive transcript diversity is generated by a relatively simple eukaryotic genome with limited splicing, and within a genetically homogeneous population of cells. Our findings have implications for genome compaction, evolution and phenotypic diversity between single cells. These data also indicate that isoform diversity as well as RNA abundance should be considered when assessing the functional repertoire of genomes.
